Building a recurrent neural network to predict time-series data with Keras in Python
Recurrent neural networks and their variants are helpful for extracting information from time series. Here’s an example using sample data to get up and running.
I found the following other websites helpful in reading up on code examples:
- https://machinelearningmastery.com/multi-step-time-series-forecasting-long-short-term-memory-networks-python/
- https://github.com/rstudio/keras/blob/master/vignettes/examples/lstm_benchmark.py
- https://github.com/jaungiers/LSTM-Neural-Network-for-Time-Series-Prediction/blob/master/lstm.py
# setup
import numpy as np
import pandas as pd
import math
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Dropout, SimpleRNN
from keras.callbacks import EarlyStopping
from sklearn.model_selection import train_test_split
# make a sample multivariable time series - not autoregressive
# generate a random walk
def random_walk(steps, scale = 1):
w = np.zeros(steps)
for x in range(1,steps):
w[x] = w[x-1] + scale * np.random.normal()
return(w)
time_steps = 5000
data = pd.DataFrame({'x' : range(time_steps),
'y' : np.arange(time_steps) ** (1/2) + random_walk(time_steps) })
data = data.assign(z = np.log(data.x+1) + 0.3 * data.y)
data_mat = np.array(data)
Take a look at the data.
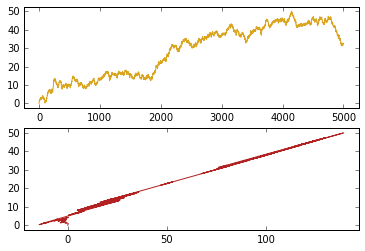
plt.subplot(2,1,1)
plt.plot(data_mat[:,0], data_mat[:,2], c = 'goldenrod')
plt.margins(0.05)
plt.subplot(2,1,2)
plt.plot(data_mat[:,1], data_mat[:,2], c = 'firebrick')
plt.margins(0.05)
plt.show()
# split into samples (sliding time windows)
samples = list()
target = list()
length = 50
# step over the 5,000 in jumps of length
for i in range(time_steps - length - 1):
# grab from i to i + length
sample = data_mat[i:i+length,:2]
outcome = data_mat[i+length+1,2]
target.append(outcome)
samples.append(sample)
# split out a test set
test_size = 1000
x_test_mat = np.dstack(samples[-test_size:])
x_test_3d_final = np.moveaxis(x_test_mat, [0, 1, 2], [1, 2, 0] )
# The RNN needs data with the format of [samples, time steps, features].
# Here, we have N samples, 50 time steps per sample, and 2 features
data_mat_stacked = np.dstack(samples[:-test_size])
data_mat_3d_final = np.moveaxis(data_mat_stacked, [0, 1, 2], [1, 2, 0] )
# and fix up the target
target_arr = np.array(target[:-test_size])
# now build the RNN
model = Sequential()
model.add(SimpleRNN(128, input_shape = (data_mat_3d_final.shape[1],
data_mat_3d_final.shape[2]), activation = 'relu'))
model.add(Dropout(0.1))
model.add(Dense(64, activation = 'relu'))
model.add(Dropout(0.1))
model.add(Dense(16, activation = 'relu'))
model.add(Dropout(0.1))
model.add(Dense(1, activation='linear'))
# monitor validation progress
early = EarlyStopping(monitor = "val_loss", mode = "min", patience = 7)
callbacks_list = [early]
model.compile(loss = 'mean_squared_error',
optimizer = 'adam',
metrics = ['mse'])
# and train the model
history = model.fit(data_mat_3d_final, target_arr,
epochs=50, batch_size=25, verbose=2,
validation_split = 0.20,
callbacks = callbacks_list)
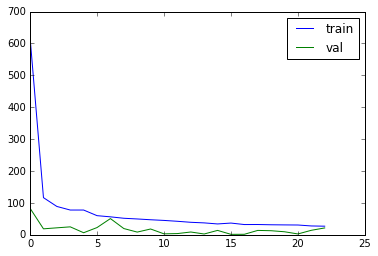
# plot history
plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='val')
plt.legend()
plt.show()
# get predictions
train_predictions = model.predict(data_mat_3d_final)
test_predictions = model.predict(x_test_3d_final)
# plot predictions vs actual
plt.plot(data['x'],
data['z'], c = 'goldenrod', label = 'data')
plt.plot(data.iloc[(length+1):-test_size]['x'],
train_predictions, c = 'navy', label = 'train')
plt.plot(data.iloc[-test_size:]['x'],
test_predictions, c = 'firebrick', label = 'test')
plt.legend(loc='best')
plt.show()
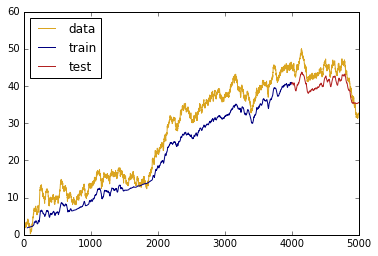
Not bad!