Rcpp is smoking fast for agent based models in data frames
Here’s the blog post originally posted on babelgraph.org on July 11, 2012. Thanks to Hadley Wickham for referencing some of content here, and apologies for the broken URL. NB. The original C++ code didn’t seem to compile on my computer today. It required a small tweak with NumericVector::create – see below.
In particular, R data frames provide a simple framework for representing large cohorts of agents in stochastic epidemiological models, such as those representing disease transmission. This approach is much easier and likely faster than trying to implement cohorts of R objects. In this post we’ll explore a simple agent-based model, and then benchmark a few different approaches to iterating through the cohort. Rcpp outperforms all of them by a few orders of magnitude. Priceless.
Case
Let’s say we are trying to predict the probability of someone choosing to receive a vaccination in a given year. The decision will be based on their age (age), gender (female), and whether or not they were infected with the virus last year (ily). Let’s make up some data:
# construct a cohort
N <- 1000 # cohort size
cohort <- data.frame(age=rnorm(N,mean=50,sd=10),
female=sample(c(0,1),size=N,replace=TRUE),
ily=sample(c(0,1),size=N,prob=c(0.8,0.2),
replace=TRUE))
The probability of choosing to be vaccinated will be given by the following function:
vaccinate <- function( age, female, ily) {
# this is based on some pretend regression equation
p <- 0.25 + 0.3 * 1/(1-exp(0.04 * age)) + 0.1 *ily
# use some logic
p <- p * ifelse(female, 1.25 , 0.75)
# boundary checking
p <- max(0,p); p <- min(1,p)
p
}
Try some iterators: for loop, apply, ddply
Let’s create a testable function for each strategy. The objective is to take a cohort data frame as input, calculate the vaccination probability for each member of the cohort, then return a data frame with the cohort data plus a new column for the vaccination probability (p).
# The traditional for loop
# Some would say is always a no-no in R...
do_forloop <- function(df) {
v_prob <- vector(length=nrow(df),mode="numeric")
for (x in 1:nrow(df)) {
v_prob[x] <- vaccinate(df$age[x],
df$female[x],df$ily[x])
}
data.frame(cbind(df,p=v_prob))
}
# The apply approach
do_apply <- function(df) {
v_prob <- apply(df, 1, function(x) vaccinate(x[1],x[2],x[3]))
data.frame(cbind(df,p=v_prob))
}
# ddply approach
library(plyr)
do_plyr <- function (df) {
v_prob <- ddply (df, names(df), function(x)
vaccinate(x$age,x$female,x$ily))
data.frame(cbind(df,p=v_prob$V1))
}
Enter Rcpp
Now rewrite the test using a traditional for-loop in C++ including a helper function to calculate the vaccination probability. I use the inline library so I can embed the C++ directly in the R script, thus obviating additional .cpp or .h files. Self-contained code is nice.
# create an R function built on C++ code
library(Rcpp)
# required for inline Rcpp calls
library(inline)
# write the C++ code
do_rcpp_src <- '
// get data from the input data frame
Rcpp::DataFrame cohort(the_cohort);
// now extract columns by name from
// the data fame into C++ vectors
std::vector<double> age_v =
Rcpp::as< std::vector<double> >(cohort["age"]);
std::vector<int> female_v =
Rcpp::as< std::vector<int> >(cohort["female"]);
std::vector<int> ily_v =
Rcpp::as< std::vector<int> >(cohort["ily"]);
// create a new variable v_prob for export
std::vector<double> v_prob (ily_v.size());
// iterate over data frame to calculate v_prob
for (int i = 0; i < v_prob.size() ; i++) {
v_prob[i] =
vaccinate_cxx(age_v[i],female_v[i],ily_v[i]);
}
// export the old with the new in a combined data frame
return Rcpp::DataFrame::create( Named("age")= age_v,
Named("female") = female_v,
Named("ily") = ily_v,
Named("p") = v_prob);
'
# write the helper function also in C++
# Note small change here from original to include Rcpp:NumericVector::create
# for use with min and max
vaccinate_cxx_src <- '
double vaccinate_cxx (double age, int female, int ily){
// this is based on some pretend regression equation
double p = 0.25 + 0.3 * 1/(1-exp(0.004*age)) + 0.1 *ily;
// use some logic
p = p * (female ? 1.25 : 0.75);
// boundary checking
p = max(Rcpp::NumericVector::create(0,p));
p = min(Rcpp::NumericVector::create(1,p));
return(p);
}
'
# create an R function to call the C++ code
do_rcpp <- cxxfunction(signature(the_cohort="data.frame"),
do_rcpp_src, plugin="Rcpp",
includes=c('#include <cmath>',
vaccinate_cxx_src))
May the best function win
# benchmarking
library(rbenchmark)
bm_results <- benchmark(do_forloop(cohort),
do_apply(cohort),
do_plyr(cohort),
do_rcpp(cohort),
replications=1000)
library(lattice)
strategy <- with(bm_results, reorder(test,relative))
barchart(relative ~ strategy, bm_results,
ylab='Relative performance',
xlab='Strategy',
main='Performance of iteration strategies over data frames in R',
col='firebrick',scales=list(x=list(cex=1.2)))
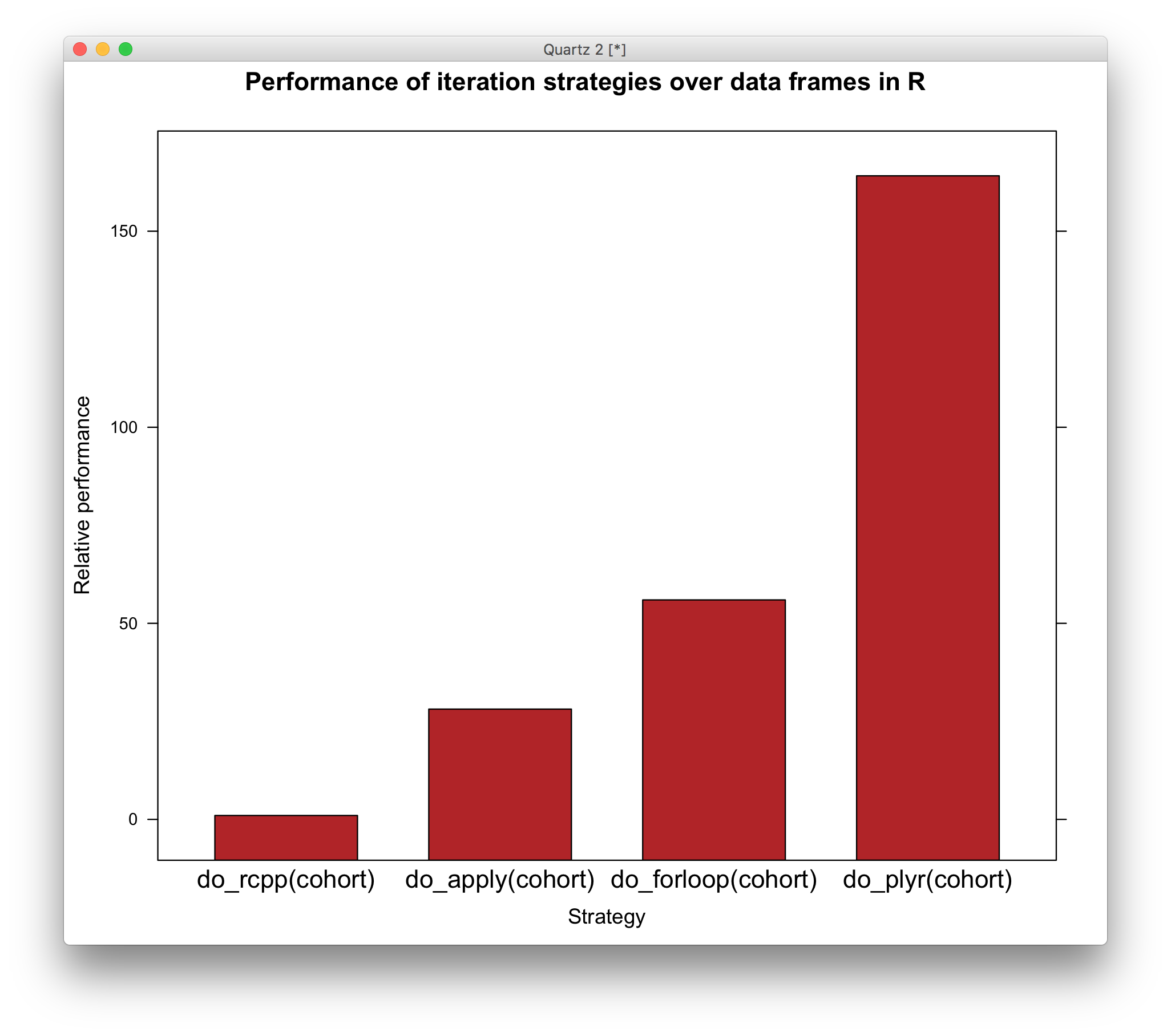
Ree – donc – u – lous.